Orthologues¶
Do(es) my gene(s) have orthologues in Human, mouse or rat?¶
- Open the following template:
- Select the gene or gene list you wish to run the search on.
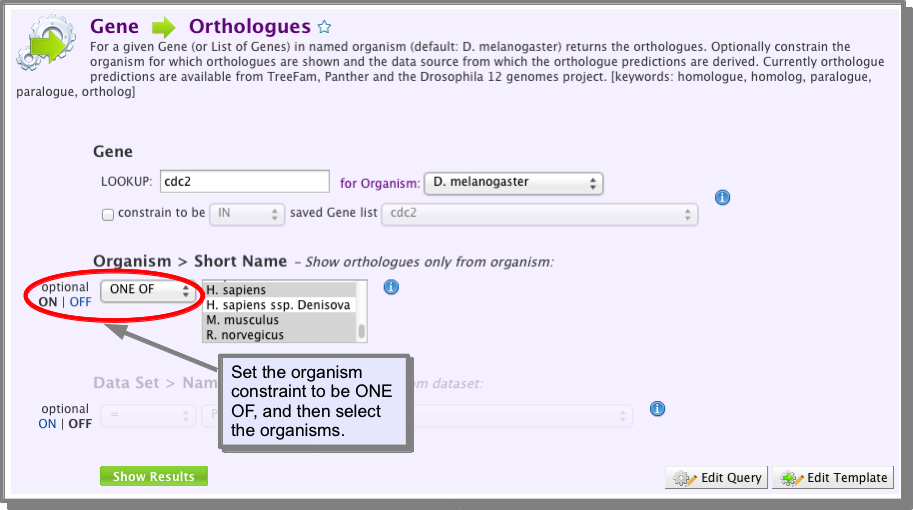
- Turn on the Organism Short Name constraint and select ONE OF and then select Human, Mouse and Rat (see image below).
- Run the template.
Does my gene(s) have a human orthologue associated with a disease?¶
In FlyMine we load disease and associated human gene data from OMIM . This data can be searched with orthologous fly genes using the following template:
Gene → Human disease gene and OMIM disease
You can also search genes associated with a specific disease. Two template searches are available, one that just returns Human genes and one that returns the Human genes and their orthologues either in all organisms or a specified organism:
- Return just the human genes:
- Return the Human gene and orthologues:
How do I export the sequences of an orthologous pair to galaxy to do an alignment?¶
- Run a template search to bring back the sequence of the gene and orthologous gene:
- Download the sequence to galaxy. You need to do this in two steps - once for the gene sequence and then for the orthologue sequence (you can’t download two sequences at once to galaxy). At this point, you need to ensure you have only one row of sequence as all rows will be downloaded. This can be achieved by further filtering your results (see Apply a filter to your results).
- In the download box, select FASTA sequence from the format options.
- Selecting the fasta option, provides a nodes option, where you can select which sequence column you are exporting.
- Under the output option you can optionally extend the sequence
- Under the destination option select Send to galaxy for analysis.
(see galaxy for more details).
- In galaxy you will need to cat your two sequence files. You can use the Concatenate datasets program under Text Manipulation in the menu bar.
- You can then use the ClustalW program in galaxy (available under Mulitple Alignments in the menu bar) to align your sequences.